Distributed representation: phenotypes and genes as inputs and features
In reading the introduction to the Deep Learning book by Ian Goodfellow, Yoshua Bengio and Aaron Courville, I came across the concept “distributed representation.” This idea struck me as parallel to the depiction of genetics in Richard Dawkins’s 1976 book The Selfish Gene.
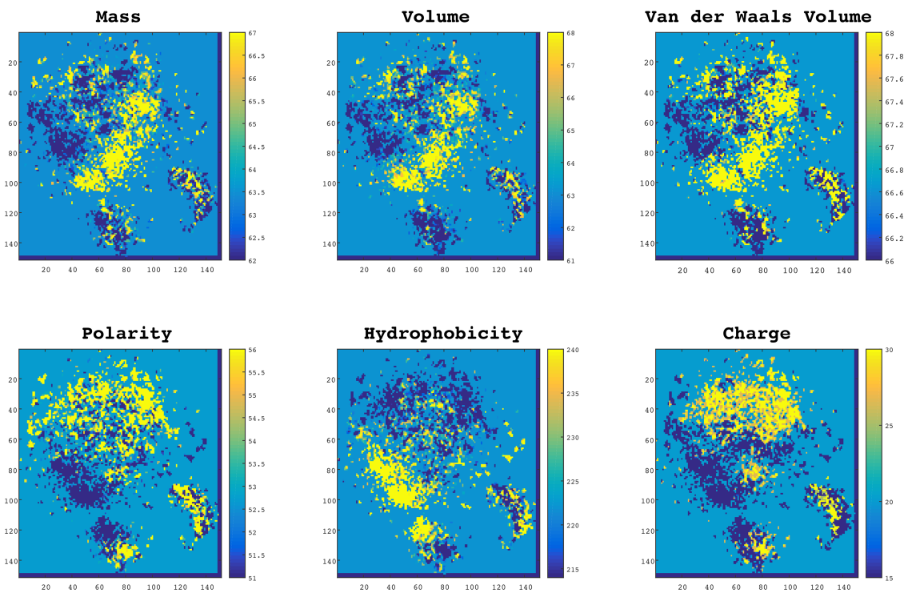
Figure 2 from: Asgari E, Mofrad MRK (2015) ContinuousDistributed Representation of Biological Sequencesfor Deep Proteomics and Genomics. PLoS ONE 10 (11): e0141287.doi:10.1371/journal.pone.0141287" class="mt-image-none" height="602
Distributed representation apparently arose during the “connectionism movement” in the 1980s, and means:
- Each input to a system should be represented by many features.
- Each feature should be involved in the representation of many possible inputs.
The given example is how distributed representation allows modeling nine vehicles of three different types, three colors each, using only six neurons: one for each color and one for each vehicle type. This is better than nine neurons each dedicated to a specific type-color combination. It’s easy to see how this is much improved, especially with larger numbers of inputs (or features? – I’m a little fuzzy still).
Throughout the introduction, the authors point out how deep learning modeling parallels biological neurons and how the brain works, but they emphasize deep learning marches on with little input from neurology. The current lack of investigative tools into brain function undoubtedly plays a role here.
Another parallel that compelled me to write this down for later reflection is the description of distributed representation matches my understanding of genes and phenotypes. I don’t think this is anything new, for we’ve known for a while some genes have multiple effects and some effects depend on multiple genes, and there’s much interdependence.
It’s interesting the structure of genes and their expressed phenotypes is similar to the structure of neurons and their computed outputs. Perhaps the general computation algorithms behind neurons are also at work in our DNA, though we don’t recognize genes as performing computation. But what else could it be? DNA sequences are clearly data. They just happen to be data that are also molecules that happen to spontaneously cause other molecules to build proteins and more that leads to a biological end result. DNA could be thought of as data that initiates its own computational evolution. It builds a computer (proteins, cells, tissues, a whole body) that does work in the universe and ends up creating new DNA, generally through reproduction, perpetuating the cycle.
So, what does that mean?
Well, my knowledge in all these areas is limited, but I want to learn more! Even though billions of years of evolution resulted in a system where the data (DNA) computes on itself, and we often find nature’s solutions are optimal, I doubt the ultimate machine learning situation would involve data creating its own computational framework in quite the same way. But who knows. This all does have me thinking about the idea the universe itself might be a computer, which I think I first read about in Ray Kurzweil’s The Singularity is Near. If all of life is based on DNA, and evolution could be looked at as DNA computation, then all of life could be a computer. I think the universe-computer idea was more along the lines of physics and quantum particles, but maybe that just means biology is a bunch of virtual machines running on a quantum computer that is the universe. (I swear I am totally sober right now.)
I did a quick search for "distributed representation" genetics to see if this is some well discussed area, but those results were overrun with information about genetic computation, which is another super fascinating area I hope to explore some day. I did find a slightly more closely related and recently published paper, “Continuous Distributed Representation of Biological Sequences for Deep Proteomics and Genomics” by Ehsaneddin Asgari and Mohammad R. K. Mofrad of University of California, Berkeley. The paper describes using deep learning to analyze protein sequences, and their “results indicate that by only providing sequence data for various
proteins into this model, accurate information about protein structure can be determined.” It seems pretty cool, but it doesn’t seem to discuss “genes as features,” as I was hoping to find.
Stuff like this makes me question what I’m doing with my life. Every time I read something in any branch of science or computing, I find myself asking questions that seem to require doctoral level research to answer. But even if I did pursue more school, I can’t do it in all directions at once!